Implementation and evaluation of an efficient dynamic data structure for DeBruijn graphs
The de Bruijn graph is a data structure which plays an important role in second-generation sequencing applications, which tend to yield a large number of short sequence fragments . It was first introduced by for de novo assembly of DNA sequences.
To get a De Bruijn graph, one should:
1.choose an appropriate value k.
2.For each k-mer that exists in any sequence create an edge with one vertex labed as the prefix and one vertex labeled as the suffix.
3.Glue all vertices that have the same label.
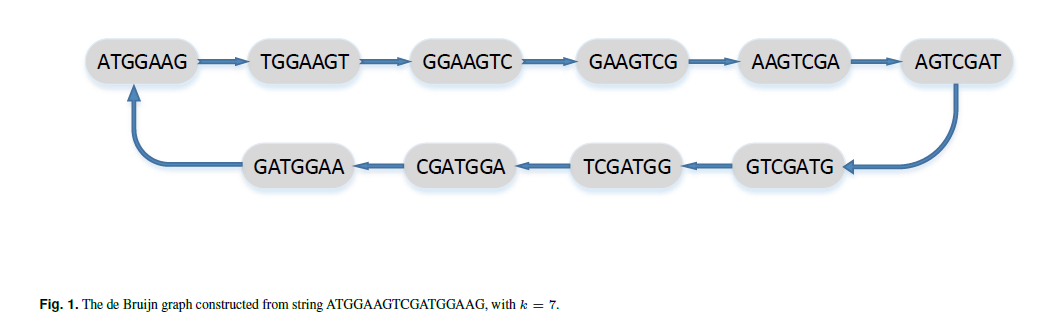
Given an alphabet set Σ of σ symbols and a set of sequencesS := {S1,S2,…,St}, the de Bruijn graph of order k onS is defined as follows. Let Mk denote the sets of k-mers (sequence of length k)thatoccurassubsequenceinSi forsomei.Adirectedgraphwithvertex set Mk is created, with directed edge e connecting node s ∈ Mk to node r ∈ Mk ifandonlythereexistsasubsequenceofsomeSi oflengthk+1 with s as a prefix and r as a suffix.
However, De Bruijn graph has it own problems in practice
-
Using de Bruijn graph in practice is the high memory occupation for certain organisms
-
human genome encoded in a de Bruijn graph with a k-mer size of 27 requires 15GB to store the node sequences
-
Further, many de novo assemblers require that the graph have a representation in-memory,and therefore space-efficiency is vital
Therefore, For our project, we will implement and evaluate the data structure described in Belazzougui’s paper to store dynamic DeBruijn graph, which allow graphs greatly reduced amount of memory